Relevant Publications
Westenberg, D.J. 2002 Evidence for AHL autoinducer production by the soybean symbiont Bradyrhizobium japonicum In Finan et al (eds.), Nitrogen Fixation: Global Perspectives. CABI Publishing, Oxford University Press, Cary, NC
Elrod, Jason C., “Identification and characterization of acyl-homoserine lactone signaling molecules produced by varying strains of Bradyrhizobium japonicum” (2004). Masters Theses. 3653.
Elrod, J. and D.J. Westenberg. Identification and characterization of acyl-homoserine lactone signaling molecules produced by strains of Bradyrhizobium japonicum (in revision)
One of the major challenges to the practical utilization of improved commercial inoculant strains is competitiveness and persistence of the inoculant strain. Competition from less efficient native strains and decreased survival in the soil make inoculant strains appear less practical and has led to a decline in the use of symbiotic nitrogen fixation as a source of plant nitrogen. One major area of investigation into the competitiveness of inoculant strains is the specific interactions between the plant and the bacteria. However, less attention has been paid to the interaction between bacteria, specifically bacterium-bacterium communication which should play a role in coordinating events in the establishment of the symbiosis. Knowledge of the “language” of bacterial communication is likely to have a significant impact on the development of commercial inoculant strains, particularly with respect to survival and competition with native strains.
Bacterium-bacterium signaling in bacteroid development. The precise sequence of events necessary for the establishment of a symbiosis and the profound changes between free-living rhizobia and bacteroids might be best viewed as a type of bacterial development. During the formation of the rhizobium/legume symbiosis, the bacterial partner is transformed from a free-living organism to an intracellular symbiont (bacteroid) that is capable of nitrogen fixation and profoundly different than the free living bacterium. The free-living bacterium must attach to the roots of the host and induce formation of nodules by the host within which nitrogen fixation takes place. In the transition from free-living to symbiotic bacterium, expression of numerous bacterial genes must be modified in response to changes in the environment. For many symbiotic genes, the signal controlling gene expression is oxygen with several bacteroid genes expressed only under oxygen-limited conditions. However, generation of an oxygen-limited environment is a fairly late event in the development of the symbiosis.
Very little is known about signals that control expression of gene products involved in the early events in the establishment of the symbiosis. What genes are regulated as rhizobia accumulate and multiply in the rhizosphere? What genes are regulated as they pass through the infection thread? What genes are regulated as the rhizobia develop from free-living cells with the goal of multiplying and searching for a symbiotic partner to bacteroids with the goal of not-growing but fixing-nitrogen instead. Because rhizobia accumulate to relatively high density around the root hair and bacteroid development occurs within the confined spaces of the infection thread and symbiosomes, genes involved in these early events are all excellent candidates for density-dependent regulation.
Several laboratories have described density-dependent phenomena in the rhizobia that are consistent with early events in development of the symbiosis – regulation of rhizosphere associated genes as the rhizobia accumulate in the rhizosphere, regulation of nodulation genes which are needed outside the host but no longer needed once inside the infection thread and growth inhibition as the rhizobia develop into bacteroids that no longer need to divide. The observation that stationary phase cultures of rhizobia capable of free-living nitrogen fixation contain non-growing cells may reflect a quorum sensing phenomenon leading to cessation of growth as the cells think they are within a nodule.
Autoinducers and their role in quorum sensing. Quorum sensing was first described in the bioluminescent bacterium Vibrio fischeri in which luminescence is expressed at high cell densities such as those found in the light organs of bioluminescent fishes. In a quorum sensing regulatory system, the bacterium produces an autoinducer molecule (AI) that is secreted to the surrounding medium. Once the AI reaches a high concentration, it interacts with a regulatory protein that modulates gene expression. In Gram-negative bacteria, two types of AIs have been observed (AI-1 and AI-2). AI-1 molecules are N-acyl-homoserine lactones (AHL) and AI-2 is a unique furanosyl borate diester. The AI-1 regulatory system consists of two structural genes – luxI that encodes the AI-1 synthase and luxR that encodes the AI-1 response regulator. LuxI and LuxR homologues are present in a wide variety of gram-negative bacteria and control numerous processes ranging from virulence genes to biofilm formation. The gene responsible for AI-2 production (luxS) is highly conserved across many species and the ability of AI-2 from a diverse group of species to regulate gene expression in other bacterial species indicates that it may have a role in inter-species communication as opposed to intra-species communication typical of AI-1 autoinducers. The AI-2 system is particular interesting because it has been correlated with pathogenicity of several organisms. With respect to symbiosis multiple signaling systems may be important in a complex community structure such as the rhizosphere where bacterial species need to coordinate their activities with bacteria of the same species as well as a host of other bacterial species. Our laboratory is screening B. japonicum culture supernatants for the presence of AI-2 signal molecules using a reporter strain but we have not detected AI-2 in B. japonucum.
Our objectives are to characterize the role of bacterium-bacterium signaling in survivability, competitiveness and nitrogen fixation of inoculant strains. One of the major drawbacks to commercial inoculant strains is the inability to compete with native strains that are less efficient at nitrogen fixation. Factors that may influence this competitiveness include: survivability of the inoculant, competition with other bacteria for binding sites on the surface of the root and appropriate bacterium-bacterium signaling to monitor the infection process. Loh et al have shown that nod genes are down-regulated by bradyoxetin, which is produced at high cell densities, such as those attained during the cultivation of inoculant strains. This demonstrates that density-dependent phenomena can influence nodulation. They have also demonstrated that mutants that are unable to produce bradyoxetin can out compete wild-type strains for nodulation, however, the resulting constitutive expression of nod genes in the nodule result in a nitrogen fixation deficiency. Our laboratory has recently initiated a new research effort to examine quorum-sensing phenomena in B. japonicum and currently has funding from the Missouri Research Board and US Department of Agriculture to characterize the AHL-like signal molecules produced by B. japonicum strain 61A227. We are also interested in determining if a correlation exists between the competitiveness of various inoculant strains and AHL production. We are currently cloning the genes likely to be responsible for AHL production and gene regulation in B. japonicum. These cloned genes will be used to make strains unable to produce or respond to these signal molecules. This project is critical for determining the role of bacterium-bacterium signaling in competitiveness and survivability. In addition, the products of this project will provide the tools to begin dissecting the regulatory networks controlling the transition from free-living bacterium to symbiotic bacteroid.
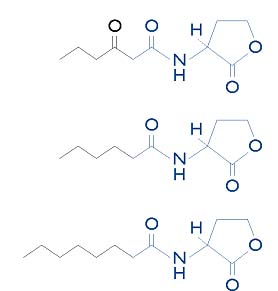
AHLs have been detected in Rhizobium leguminosarum, Rhizobium etli, and Rhizobium meliloti and in many cases, multiple AHL molecules are detected. In R. leguminosarum, AHLs are required for activation of the rhiABC operon (a set of rhizosphere-expressed genes), raiIR and cinIR genes and is involved in root nodulation and growth inhibition. Until recently, AHL autoinducers had not been detected in B. japonicum the symbiont of soybean, a major agricultural crop. In the next section we present evidence for the production of an N-acyl homoserine lactone AI by several strains of B. japonicum.
Using the NTL4/pZLR4 indicator strain described by Piper et al we screened twenty-three strains of B. japonicum for production of AHL autoinducer(s). Agrobacterium strain NTL4 does not produce AHL and the plasmid pZLR4 contains the Agrobacterium traR gene (required for AHL dependent gene regulation) and a gene fusion between the Agrobacterium traG gene and the lacZ gene from Escherichia coli. TraR induction of the traG::lacZ gene fusion requires AHL; therefore, this strain does not express the traG::lacZ fusion unless provided exogenous AHL. Culture supernatants from AHL producing strains are capable of inducing traG::lacZ when added to cultures of the NTL4/pZLR4 indicator strain.